R Playbook: Introduction to Mixed-Models
I just started reading more about mixed-models (multilevel/hierarchical) and will use this as a playbook. Mostly becaus I learn the best by experimenting with the data and I suggest everyone to try to do the same. So please note that this is just a published playbook – if you find it useful, great, if you find some errors, please let me know, if you find it useless, well, disregard it.
Anyway, here is our sleep study data from lme4 package:
The data represents the average reaction time per day for subjects in a sleep deprivation study. On day 0 the subjects had their normal amount of sleep. Starting that night they were restricted to 3 hours of sleep per night. The observations represent the average reaction time on a series of tests given each day to each subject.
Basically repeated measures design. Let’s see the relationship between days and reaction time.
gg <- ggplot(sleepstudy, aes(x = Days, y = Reaction))
gg <- gg + geom_point(color = "blue", alpha = 0.7)
gg <- gg + geom_smooth(method = "lm", color = "black")
gg <- gg + theme_bw()
gg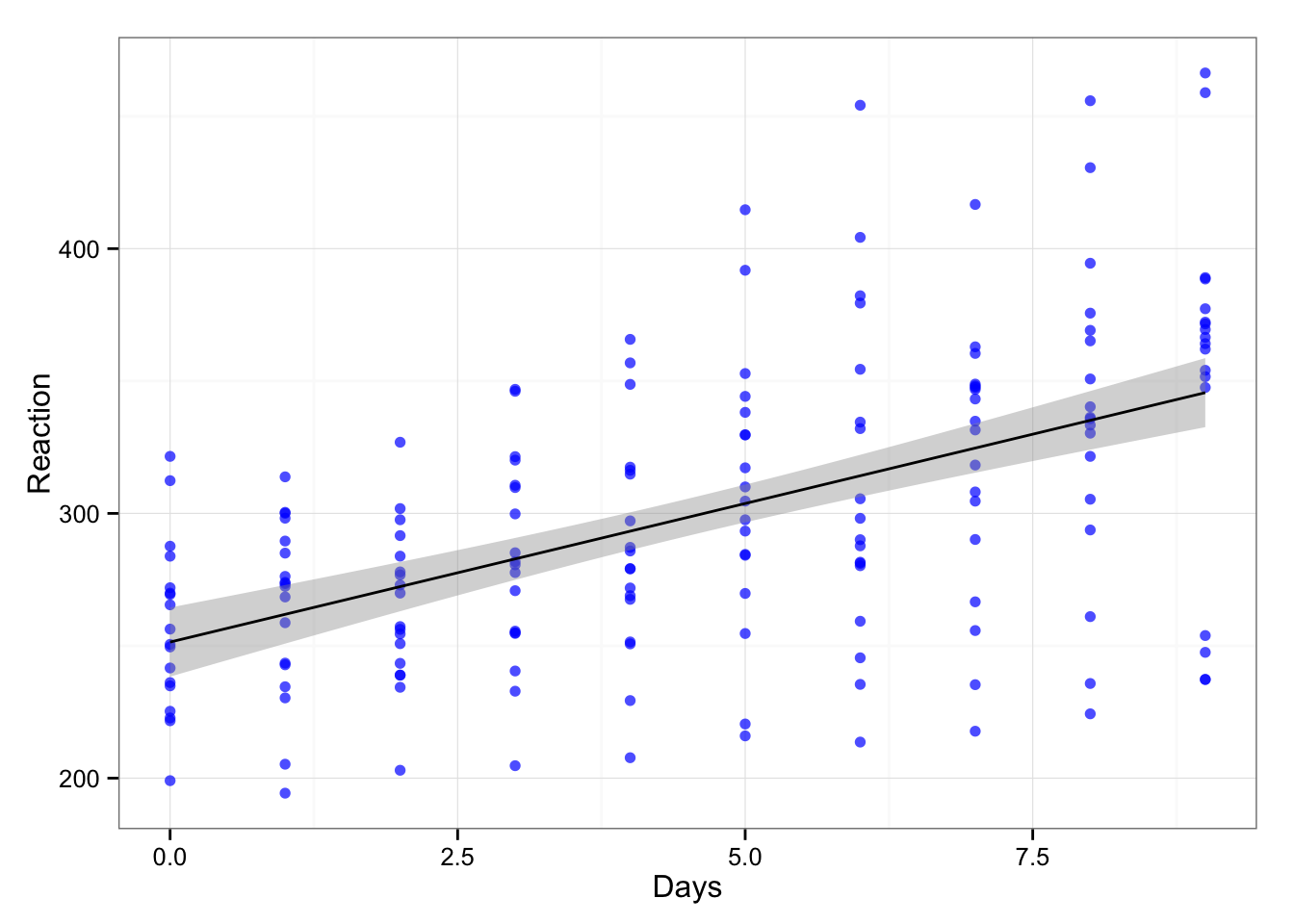
If we create simple linear model we get the following
model1 <- lm(Reaction ~ Days, sleepstudy)
summary(model1)##
## Call:
## lm(formula = Reaction ~ Days, data = sleepstudy)
##
## Residuals:
## Min 1Q Median 3Q Max
## -110.848 -27.483 1.546 26.142 139.953
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 251.405 6.610 38.033 < 2e-16 ***
## Days 10.467 1.238 8.454 9.89e-15 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 47.71 on 178 degrees of freedom
## Multiple R-squared: 0.2865, Adjusted R-squared: 0.2825
## F-statistic: 71.46 on 1 and 178 DF, p-value: 9.894e-15And our coefficients are the following:
coef(model1)## (Intercept) Days
## 251.40510 10.46729What we learned from the model is that days with sleep deprivations are negativelly affecting reaction time. The 95% CIs are:
round(confint(model1), 2)## 2.5 % 97.5 %
## (Intercept) 238.36 264.45
## Days 8.02 12.91But we included all the subjects into one bin and there is probably variation within the subjects. Let’s plot the trellis graph across the subjects:
gg <- gg + facet_wrap(~Subject)
gg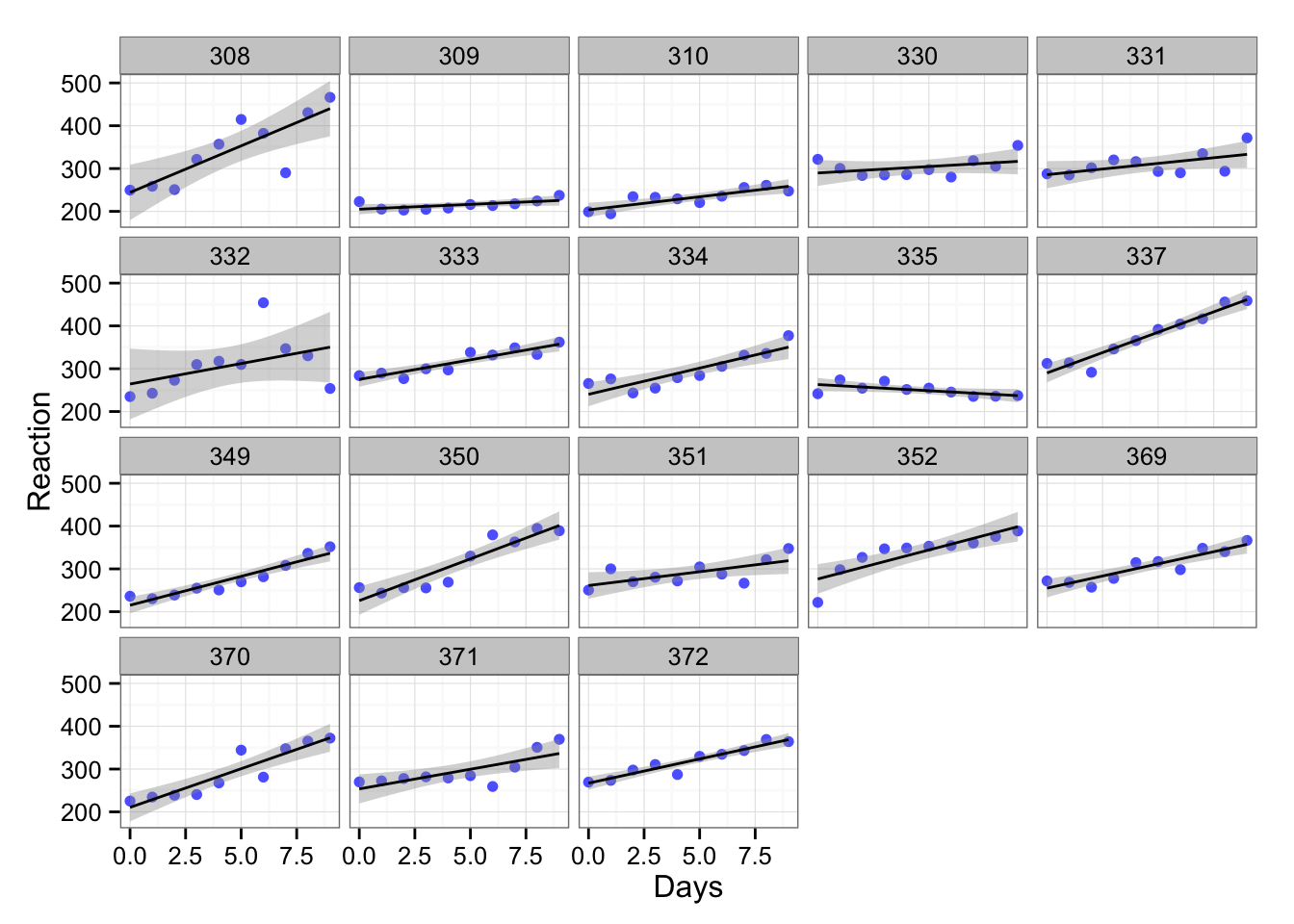
In the above graph each subject is depicted in single box (the number above is the code of the subject). As we can see the subject reaction differs. What we can do is to create linear model for each subject. Let’s do it for subject 308 and subject 335
model308 <- lm(Reaction ~ Days,
data = subset(sleepstudy, Subject == 308))
model335 <- lm(Reaction ~ Days,
data = subset(sleepstudy, Subject == 335))
summary(model308)##
## Call:
## lm(formula = Reaction ~ Days, data = subset(sleepstudy, Subject ==
## 308))
##
## Residuals:
## Min 1Q Median 3Q Max
## -106.397 -4.098 9.688 22.269 61.674
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 244.19 28.08 8.695 2.39e-05 ***
## Days 21.77 5.26 4.137 0.00326 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 47.78 on 8 degrees of freedom
## Multiple R-squared: 0.6815, Adjusted R-squared: 0.6417
## F-statistic: 17.12 on 1 and 8 DF, p-value: 0.003265summary(model335)##
## Call:
## lm(formula = Reaction ~ Days, data = subset(sleepstudy, Subject ==
## 335))
##
## Residuals:
## Min 1Q Median 3Q Max
## -21.4264 -3.8697 -0.1774 4.5403 16.4105
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 263.035 6.694 39.296 1.93e-10 ***
## Days -2.881 1.254 -2.298 0.0506 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 11.39 on 8 degrees of freedom
## Multiple R-squared: 0.3976, Adjusted R-squared: 0.3223
## F-statistic: 5.28 on 1 and 8 DF, p-value: 0.05065Subject 308 have positive coefficient (and statistically significant), where subject 335 have negative coefficeint (and nearly statistically significant).
round(confint(model308), 2)## 2.5 % 97.5 %
## (Intercept) 179.43 308.95
## Days 9.63 33.90round(confint(model335), 2)## 2.5 % 97.5 %
## (Intercept) 247.60 278.47
## Days -5.77 0.01Let’a create linear models across subjects
model2 <- lmList(Reaction ~ Days | Subject, sleepstudy)
model2.coefficients <- coef(model2)
names(model2.coefficients) <- c("Intercept", "Days.Slope")We have intercept and slope for each subject
And we can plot them:
gg <- ggplot(model2.coefficients, aes(x = Intercept, y = Days.Slope))
gg <- gg + geom_point()
gg <- gg + theme_bw()
gg <- gg + geom_smooth(method = "loess")
gg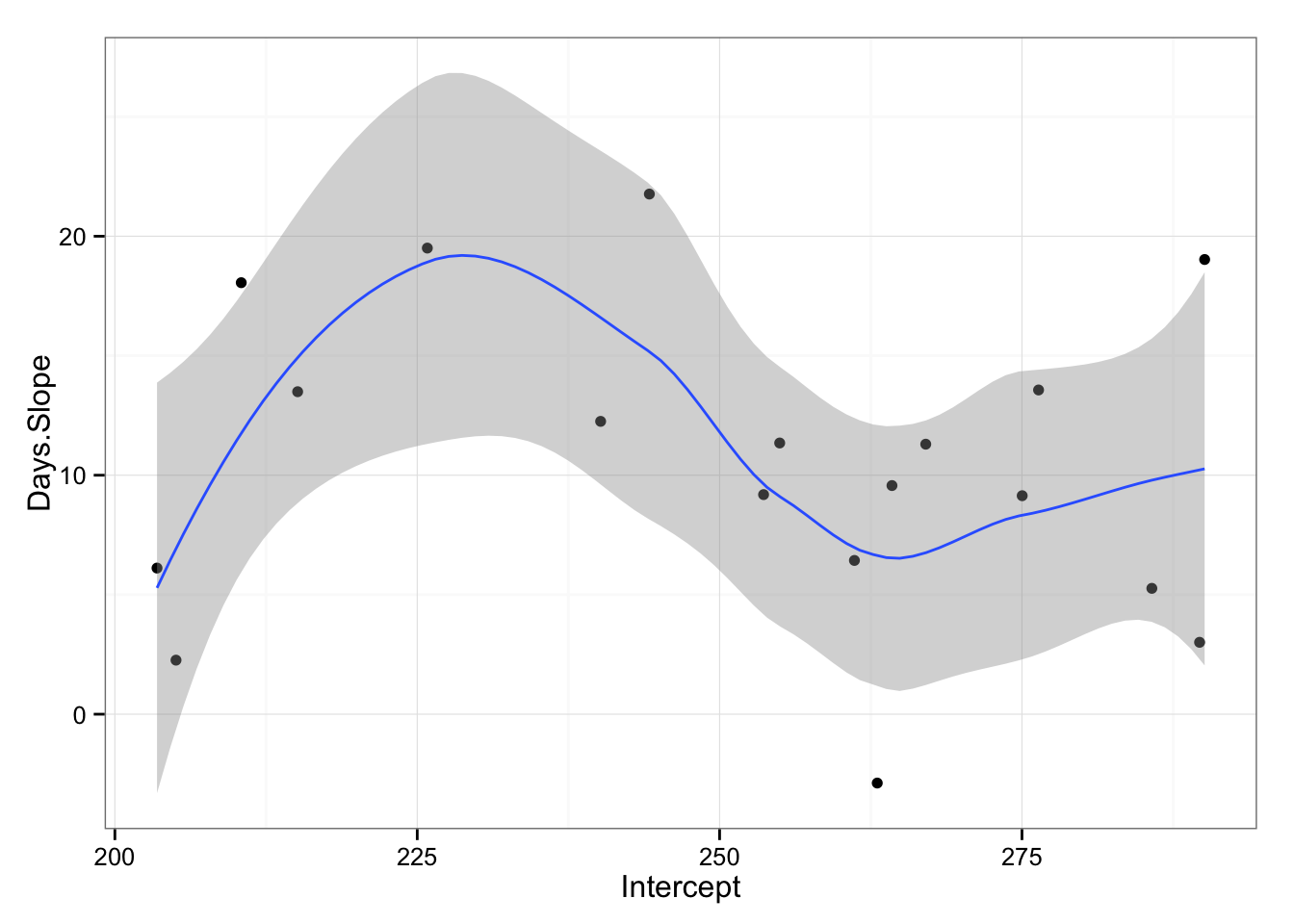
What we are interested to see from the graph is that if there is relationship from starting reaction time and slope across subjects. As can be seen from the picture it appears that there is non-linear relationship. We can also try to apply linear model (even if we can see that things are not linear) just to check for the trend.
gg <- ggplot(model2.coefficients, aes(x = Intercept, y = Days.Slope))
gg <- gg + geom_point()
gg <- gg + theme_bw()
gg <- gg + geom_smooth(method = "lm")
gg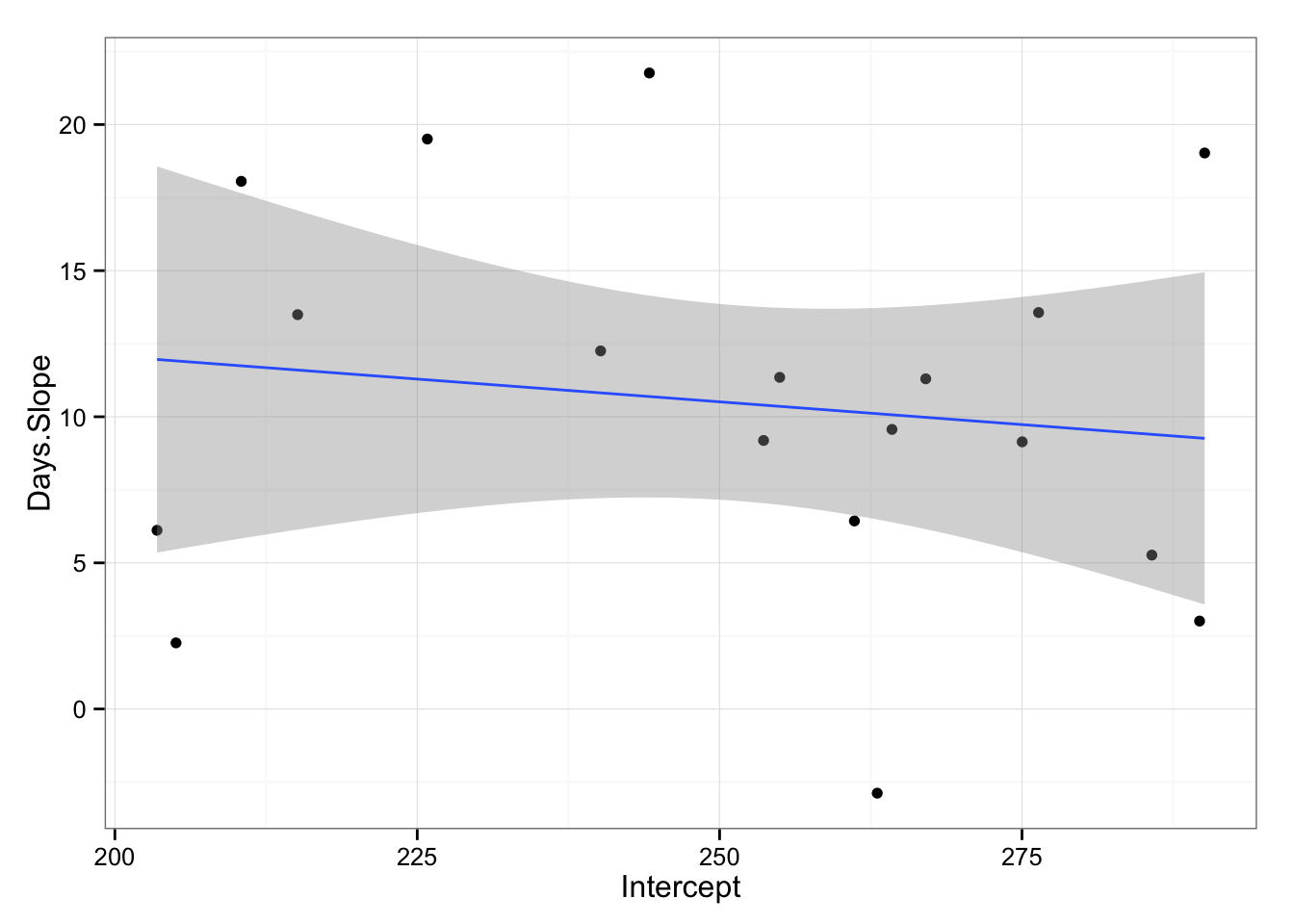
model3 <- lm(Days.Slope ~ Intercept, model2.coefficients)
summary(model3)##
## Call:
## lm(formula = Days.Slope ~ Intercept, data = model2.coefficients)
##
## Residuals:
## Min 1Q Median 3Q Max
## -12.9860 -4.0312 0.2458 3.3818 11.0727
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 18.30016 14.18878 1.290 0.215
## Intercept -0.03116 0.05609 -0.555 0.586
##
## Residual standard error: 6.696 on 16 degrees of freedom
## Multiple R-squared: 0.01892, Adjusted R-squared: -0.0424
## F-statistic: 0.3086 on 1 and 16 DF, p-value: 0.5862As can be seen the trend is that subjects with slower initial reaction time have slower slope (or decrement) on their reaction time with sleep deprivation (not statistically significant, p = 0.582). Hence, slower individuals are less affected by sleep deprivation, even if the fixed effect is that everyone is affected by sleep deprivation negativelly (increase in reaction time, p < 0.001 ).
This can be easily performed using lme4 package.
mix.model1 <- lmer(Reaction ~ Days + (Days | Subject), sleepstudy)
summary(mix.model1)## Linear mixed model fit by REML ['lmerMod']
## Formula: Reaction ~ Days + (Days | Subject)
## Data: sleepstudy
##
## REML criterion at convergence: 1743.6
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.9536 -0.4634 0.0231 0.4634 5.1793
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 612.09 24.740
## Days 35.07 5.922 0.07
## Residual 654.94 25.592
## Number of obs: 180, groups: Subject, 18
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 251.405 6.825 36.84
## Days 10.467 1.546 6.77
##
## Correlation of Fixed Effects:
## (Intr)
## Days -0.138To be honest, I can understand a little bit of the lme4 model output. Anyway, we managed to take into acount the levels (in this case individual athletes) can create a model that calculates overal effects (fixed effects) of days of sleep deprivation to reaction time, but also take into account invidual differences and effect of starting reaction time (intervcept) to the slope (or effect of sleep deprivation) for each individual.
Back to the bat cave. To be continued…..
In the mean time I am reading the following book:








Responses